cubes.piws.importer.genetics.Genetics¶
-
class
cubes.piws.importer.genetics.Genetics(session, project_name, center_name, genetics, can_read=True, can_update=True, data_filepath=None, use_store=True, piws_security_model=True)[source]¶ This class enables us to load the genetic data to CW.
-
__init__(session, project_name, center_name, genetics, can_read=True, can_update=True, data_filepath=None, use_store=True, piws_security_model=True)[source]¶ Initialize the Genetics class.
Parameters: session: Session (mandatory) :
a cubicweb session.
project_name: str (mandatory) :
the name of the project
center_name: str (mandatory) :
the center name
genetics: dict of list of dict (mandatory) :
the genetic measure description: the first dictionary contains the timepoint as keys and then a list of dictionaries with two keys (GenomicMeasures - Assessment) that contains the entities parameter descriptions.
can_read: bool (optional, default True) :
set the read permission to the imported data.
can_update: bool (optional, default True) :
set the update permission to the imported data.
data_filepath: str (optional, default None) :
the path to folder containing the current study dataset.
use_store: bool (optional, default True) :
if True use an SQLGenObjectStore, otherwise the session.
piws_security_model: bool (optional, default True) :
if True apply the PIWS security model.
Notes
Here is an example of the definition of the ‘genetics’ parameter:
genetics = { "V1": [{ "GenomicMeasures": [{ "GenomicPlatform": { "related_snps": [ u"rs325623", u"rs272569", u"rs400", u"rs1053026" ], "name": u"Illumina", "related_subjects": [ u"subject0", u"subject1", u"subject2", u"subject3", u"subject4", u"subject5", u"subject6", u"subject7", u"subject8", u"subject9" ] }, "GenomicMeasure": { "identifier": u"toy_V1_genetic_Illumina_raw_json", "label": u"toy_V1_genetic_Illumina_raw_json", "type": u"raw", "format": u"json" }, "FileSet": { "identifier": u"toy_V1_genetic", "name": u"raw genetic measure" }, "ExternalResources": [{ "absolute_path": True, "identifier": u"toy_V1_genetic_1", "name": u"genetic", "filepath": u"/tmp/demo/V1/genetic/genetic.json" }] }], "Assessment": { "identifier": u"toy_V1_genetic", "timepoint": u"V1"} }] }
Methods
__init__(session, project_name, center_name, ...)Initialize the Genetics class. cleanup()Method to cleanup temporary items and to commit changes. import_data()Method that import some genetic data in the db. schema(outfname[, text_font, node_text_size])Create a view of the schema described in a python structure. Attributes
assessment_relationsdevice_relationsfileset_relationsrelations-
__module__= 'cubes.piws.importer.genetics'¶
-
_create_measure(measure_struct, fset_struct, extfiles, related_subjects, subjects_eid_map, study_eid, assessment_eid, platform_eid)[source]¶ Create a genomic measure and its associated relations.
-
_create_platform(platform_struct, related_snps)[source]¶ Create a genomic platform and its associated relations.
-
import_data()[source]¶ Method that import some genetic data in the db.
Warning
This method assumes that all the subjects and groups have already been inserted in the database.
-
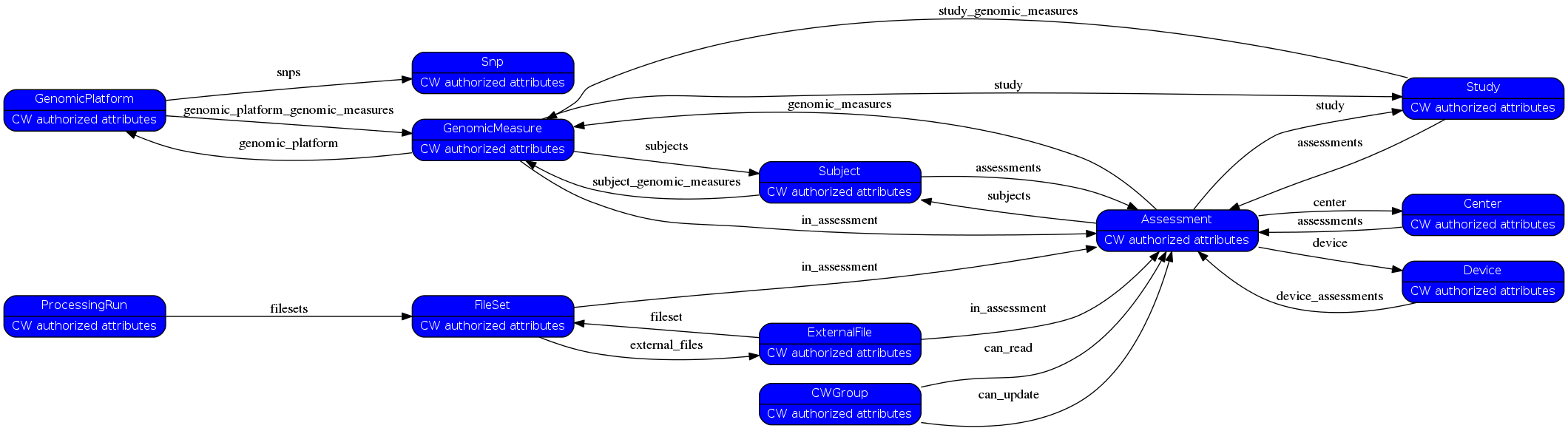
relations= [['Scan', 'filesets', 'FileSet'], ('FileSet', 'in_assessment', 'Assessment'), ('FileSet', 'external_files', 'ExternalFile'), ('ExternalFile', 'fileset', 'FileSet'), ('ExternalFile', 'in_assessment', 'Assessment'), ('Assessment', 'study', 'Study'), ('Study', 'assessments', 'Assessment'), ('Subject', 'assessments', 'Assessment'), ('Assessment', 'subjects', 'Subject'), ('Center', 'assessments', 'Assessment'), ('Assessment', 'center', 'Center'), ('CWGroup', 'can_read', 'Assessment'), ('CWGroup', 'can_update', 'Assessment'), ('Assessment', 'device', 'Device'), ('Device', 'device_assessments', 'Assessment'), ('GenomicMeasure', 'study', 'Study'), ('Study', 'study_genomic_measures', 'GenomicMeasure'), ('GenomicMeasure', 'subjects', 'Subject'), ('Subject', 'subject_genomic_measures', 'GenomicMeasure'), ('Assessment', 'genomic_measures', 'GenomicMeasure'), ('GenomicMeasure', 'in_assessment', 'Assessment'), ('GenomicMeasure', 'genomic_platform', 'GenomicPlatform'), ('GenomicPlatform', 'genomic_platform_genomic_measures', 'GenomicMeasure'), ('GenomicPlatform', 'snps', 'Snp')]¶
-