cubes.piws.importer.scans.Scans¶
-
class
cubes.piws.importer.scans.Scans(session, project_name, center_name, scans, can_read=True, can_update=False, data_filepath=None, use_store=True, piws_security_model=True)[source]¶ This class enables us to load the scan data to CW.
-
__init__(session, project_name, center_name, scans, can_read=True, can_update=False, data_filepath=None, use_store=True, piws_security_model=True)[source]¶ Initialize the Scans class.
Parameters: session: Session (mandatory) :
a cubicweb session.
project_name: str (mandatory) :
the name of the project.
center_name: str (mandatory) :
the center name.
scans: dict of list of dict (mandatory) :
the scan description: the first dictionary contains the subject name as keys and then a list of dictionaries with four keys (Scans - (Scan - TypeData - FileSet - ExternalResource - ScoreValues) - Assessment) that contains the entities parameter decriptions.
can_read: bool (optional, default True) :
set the read permission to the imported data.
can_update: bool (optional, default False) :
set the update permission to the imported data.
data_filepath: str (optional, default None) :
the path to folder containing the current study dataset.
use_store: bool (optional, default True) :
if True use an SQLGenObjectStore, otherwise the session.
piws_security_model: bool (optional, default True) :
if True apply the PIWS security model.
Notes
Here is an axemple of the definiton of the ‘scans’ parameter:
scans = { "subjects1": [{ "Assessment": { "age_of_subject": 27, "identifier": u"toy_V1_subject1", "timepoint": u"V1"}, "Scans": [{ "TypeData": { "fov_y": 0, "fov_x": 0, "voxel_res_y": 2.0, "voxel_res_x": 2.0, "voxel_res_z": 2.0, "field": "3T", "tr": 2.5, "shape_y": 2, "shape_x": 2, "shape_z": 2, "te": 0, "type": u"MRIData"}, "ExternalResources": [ { "absolute_path": True, "name": u"t1", "identifier": u"toy_V1_subject1_t1_1", "filepath": u"/tmp/demo/V1/subject1/images/t1/t1.nii.gz"}], "FileSet": { "identifier": u"toy_V1_subject1_t1", "name": u"T1"}, "Scan": { "format": u"Nifti", "label": u"T1", "identifier": u"toy_V1_subject1_t1", "type": u"MRIData"} }] }] }
Note that an optional filed can be set in order to specify the scan acquisition device.
scans = { "subjects1": [ { "Assessment": { .. }, "Device": { "ExternalResources": [{ "absolute_path": true, "filepath": "/my/path/examcard.pdf", "identifier": "EXAM_CARD_TIEMPOINT_CENTER", "name": "EXAM_CARD_TIEMPOINT_CENTER" }], "identifier": "31be53546754dc5f04ab2d9db6bed7cf", "manufacturer": "SIEMENS", "model": "Verio", "serialnum": "xxxxx", "software_version": "xxxxxx" } ... }] }
Methods
__init__(session, project_name, center_name, ...)Initialize the Scans class. cleanup()Method to cleanup temporary items and to commit changes. import_data()Method that import the scan data in the db. schema(outfname[, text_font, node_text_size])Create a view of the schema described in a python structure. Attributes
assessment_relationsdevice_relationsdtypefileset_relationshas_datarelations-
__module__= 'cubes.piws.importer.scans'¶
-
_create_scan(scan_struct, scantype_struct, fset_struct, extfiles, scores, subject_eid, study_eid, assessment_eid)[source]¶ Create a scans and its associated relations.
-
dtype= 'PETData'¶
-
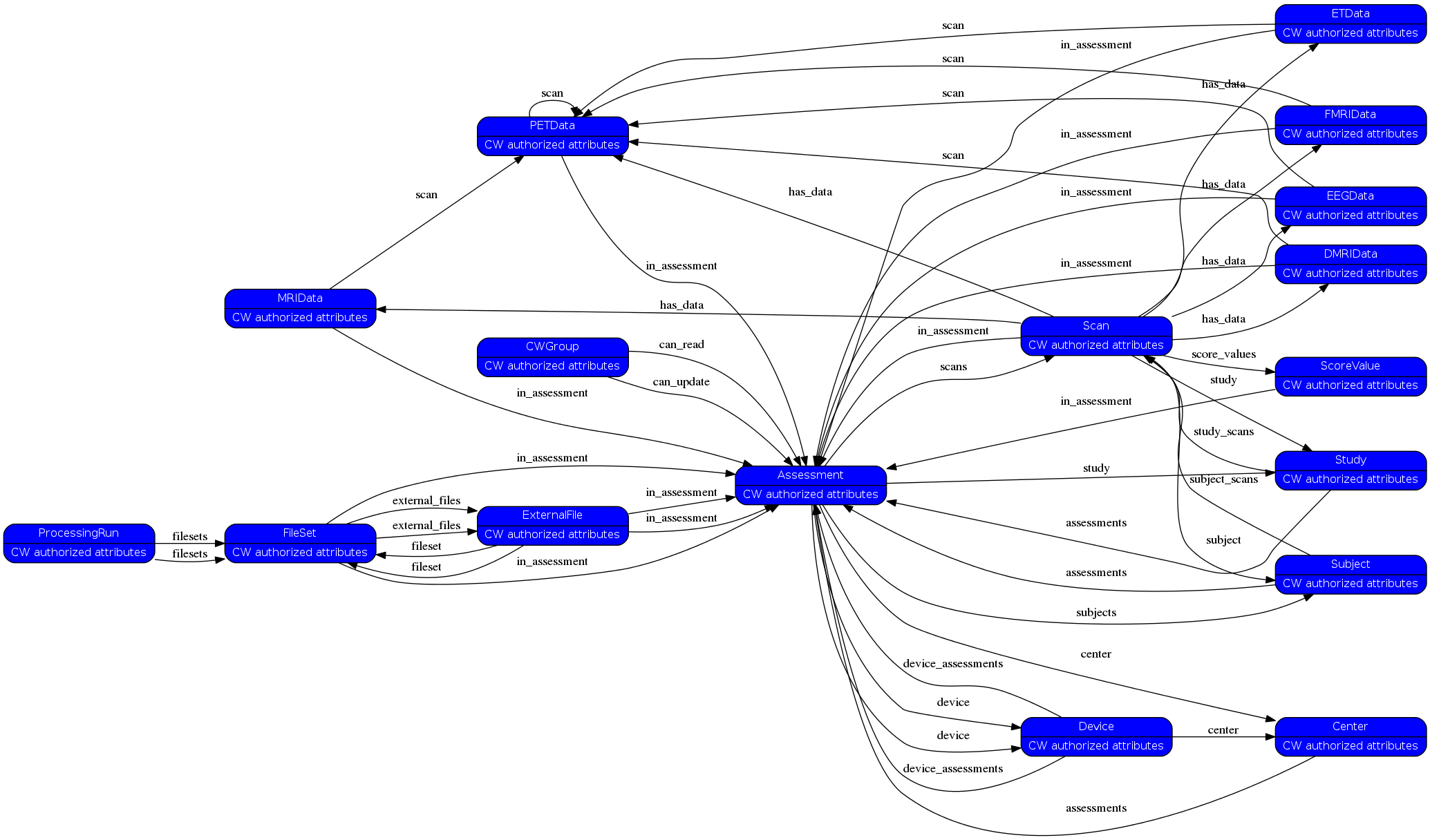
has_data= [('Scan', 'has_data', 'MRIData'), ('MRIData', 'scan', 'PETData'), ('MRIData', 'in_assessment', 'Assessment'), ('Scan', 'has_data', 'DMRIData'), ('DMRIData', 'scan', 'PETData'), ('DMRIData', 'in_assessment', 'Assessment'), ('Scan', 'has_data', 'FMRIData'), ('FMRIData', 'scan', 'PETData'), ('FMRIData', 'in_assessment', 'Assessment'), ('Scan', 'has_data', 'EEGData'), ('EEGData', 'scan', 'PETData'), ('EEGData', 'in_assessment', 'Assessment'), ('Scan', 'has_data', 'ETData'), ('ETData', 'scan', 'PETData'), ('ETData', 'in_assessment', 'Assessment'), ('Scan', 'has_data', 'PETData'), ('PETData', 'scan', 'PETData'), ('PETData', 'in_assessment', 'Assessment')]¶
-
import_data()[source]¶ Method that import the scan data in the db.
Warning
In the ‘scans’ input structure, the ‘TypeData’ item contains a special key ‘type’ corresponding to the data type. The associated value is a string representing the entity name that must be in [‘PETData’, ‘FMRIData’, ‘DMRIData’, ‘MRIData’].
Warning
This method assumes that all the subjects and groups have already been inserted in the database.
-
relations= [['Scan', 'filesets', 'FileSet'], ('FileSet', 'in_assessment', 'Assessment'), ('FileSet', 'external_files', 'ExternalFile'), ('ExternalFile', 'fileset', 'FileSet'), ('ExternalFile', 'in_assessment', 'Assessment'), ('Assessment', 'study', 'Study'), ('Study', 'assessments', 'Assessment'), ('Subject', 'assessments', 'Assessment'), ('Assessment', 'subjects', 'Subject'), ('Center', 'assessments', 'Assessment'), ('Assessment', 'center', 'Center'), ('CWGroup', 'can_read', 'Assessment'), ('CWGroup', 'can_update', 'Assessment'), ('Assessment', 'device', 'Device'), ('Device', 'device_assessments', 'Assessment'), ['Scan', 'filesets', 'FileSet'], ('FileSet', 'in_assessment', 'Assessment'), ('FileSet', 'external_files', 'ExternalFile'), ('ExternalFile', 'fileset', 'FileSet'), ('ExternalFile', 'in_assessment', 'Assessment'), ('Device', 'center', 'Center'), ('Scan', 'study', 'Study'), ('Study', 'study_scans', 'Scan'), ('Scan', 'subject', 'Subject'), ('Subject', 'subject_scans', 'Scan'), ('Assessment', 'scans', 'Scan'), ('Scan', 'in_assessment', 'Assessment'), ('Scan', 'score_values', 'ScoreValue'), ('Assessment', 'device', 'Device'), ('Device', 'device_assessments', 'Assessment'), ('ScoreValue', 'in_assessment', 'Assessment'), ('Scan', 'has_data', 'MRIData'), ('MRIData', 'scan', 'PETData'), ('MRIData', 'in_assessment', 'Assessment'), ('Scan', 'has_data', 'DMRIData'), ('DMRIData', 'scan', 'PETData'), ('DMRIData', 'in_assessment', 'Assessment'), ('Scan', 'has_data', 'FMRIData'), ('FMRIData', 'scan', 'PETData'), ('FMRIData', 'in_assessment', 'Assessment'), ('Scan', 'has_data', 'EEGData'), ('EEGData', 'scan', 'PETData'), ('EEGData', 'in_assessment', 'Assessment'), ('Scan', 'has_data', 'ETData'), ('ETData', 'scan', 'PETData'), ('ETData', 'in_assessment', 'Assessment'), ('Scan', 'has_data', 'PETData'), ('PETData', 'scan', 'PETData'), ('PETData', 'in_assessment', 'Assessment')]¶
-