Cortical brain surface and deep learning
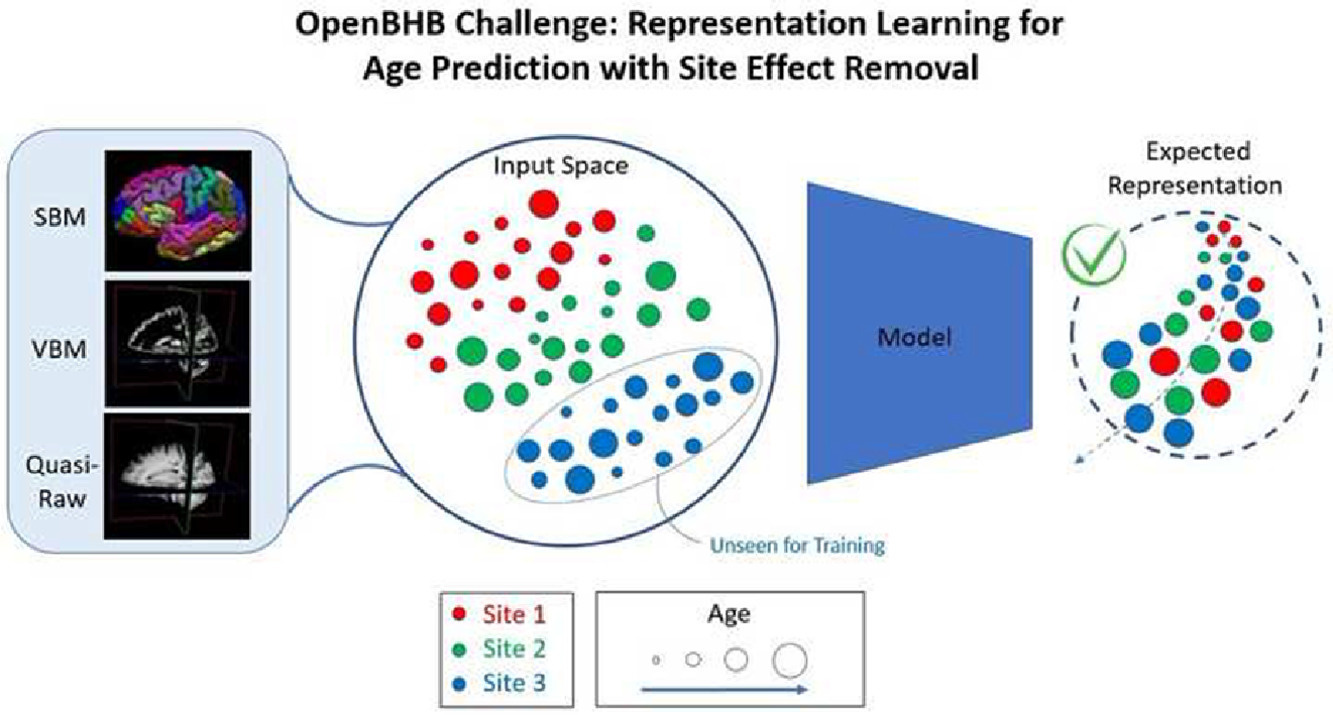
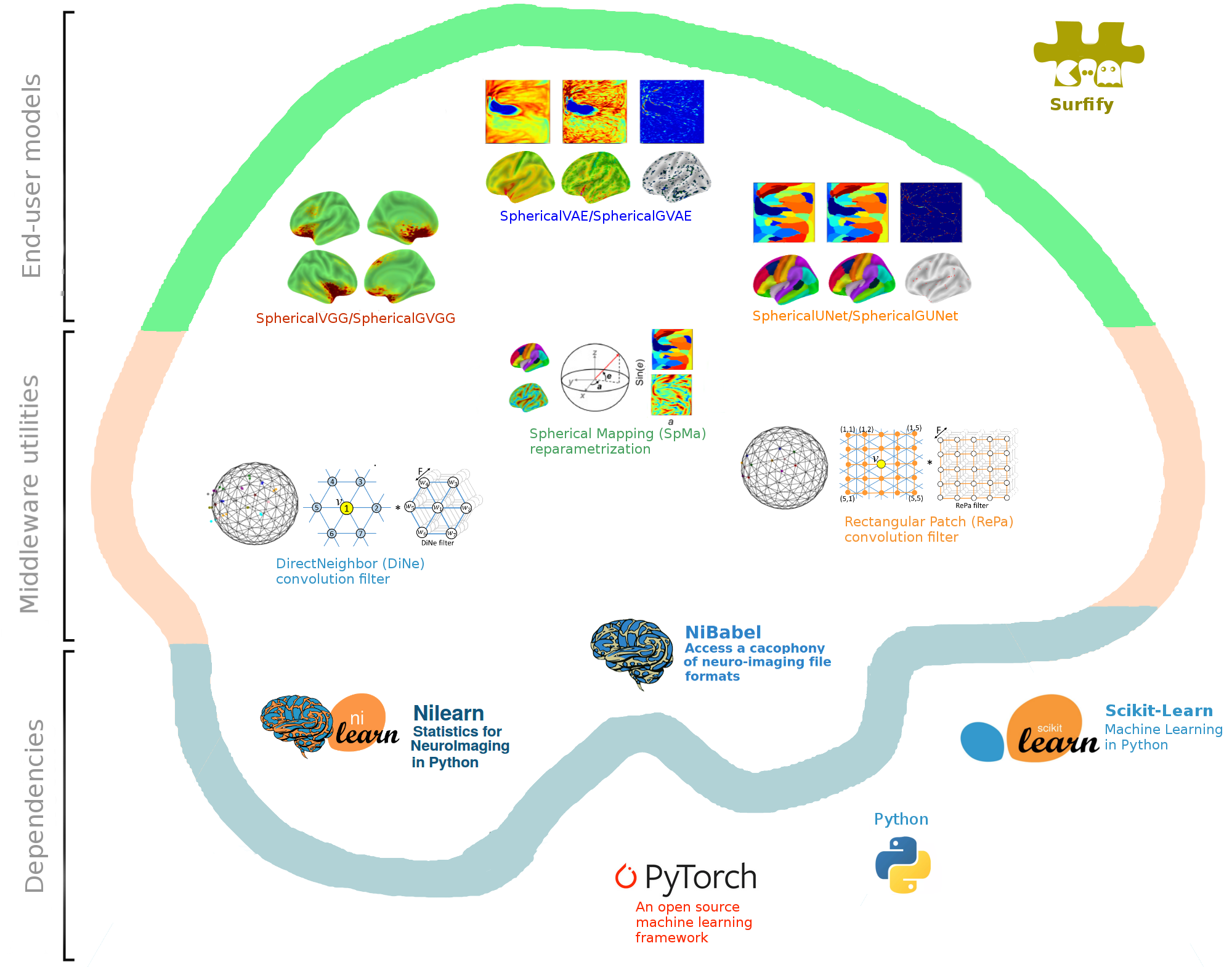
Surfify is an open source Python module that simplifies the development of neural network architectures that relies on cortical surfaces. It provides common architectures, icosahedral mesh operators, and cortical augmentations.